Kidney-specific knockout mouse modeling via conditional disruption of PBRM1 gene
Recently, a new kidney cancer-related gene PBRM1 was identified through exome sequencing in a series of primary clear cell renal carcinoma (ccRCC). Study showed that around 40% examined cases carried PBRM1 gene mutations, indicating PBRM1 is the second most important kidney cancer-related gene following VHL gene whose mutations account for approximate 70% of ccRCC cases.
Human PBRM1 gene lies on 3p21, contains 30 exons (variant 1), and encodes a protein (BAF180) of 1582 amino acides. The mouse Pbrm1 gene is located on chromosome 14, composed of 33 exons, and encodes a protein of 1704 amino acids.
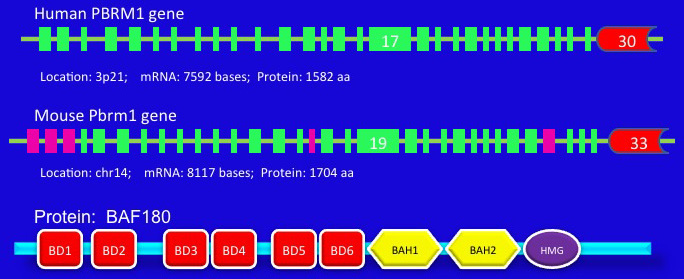
PBRM1 gene structure and corresponding protein structure.
PBRM1 protein BAF180 is a member of SWI/SNF complex (SWItch/Sucrose NonFermentable). The SWI/SNF complex is an evolutionarily conserved multi-subunit chromatin-remodelling complex, which uses the energy of ATP hydrolysis to mobilize nucleosomes and remodel chromatin. It has been found that the SWI/SNF complex (in yeast) is capable of altering the position of nucleosomes along DNA. Two mechanisms for nucleosome remodeling by SWI/SNF have been proposed. The first model contends that a unidirectional diffusion of a twist defect within the nucleosomal DNA results in a corkscrew-like propagation of DNA over the octamer surface that initiates at the DNA entry site of the nucleosome. The other is known as the "bulge" or "loop-recapture" mechanism and it involves the dissociation of DNA at the edge of the nucleosome with reassociation of DNA inside the nucleosome, forming a DNA bulge on the octamer surface. The DNA loop would then propagate across the surface of the histone octamer in a wave-like manner, resulting in the repositioning of DNA without changes in the total number of histone-DNA contacts. A recent study has provided strong evidence against the twist diffusion mechanism and has further strengthened the loop-recapture model.
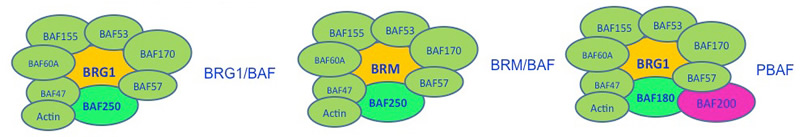
The mammalian SWI/SNF complexes mediate ATP-dependent chromatin remodeling processes that are critical for differentiation and proliferation. Not surprisingly, loss of SWI/SNF function has been associated with malignant transformation, and a substantial body of evidence indicates that several components of the SWI/SNF complexes function as tumor suppressors.
To study the biological function of PBRM1and its role in kidney tumorigenesis, we have been developing a few PBRM1 knockout mouse models by deleting the exon 2.
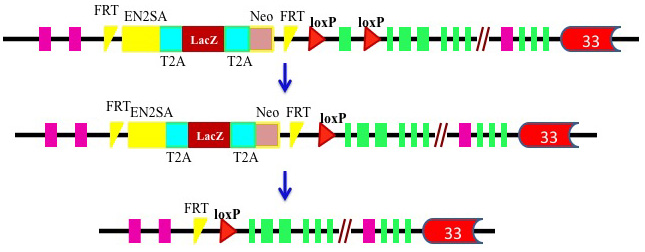
Click here to see BHD knockout mouse models